Zandile Nare, PhD | 17th June 2025
Eosinophils are granulocytic leukocytes that are produced from CD34+ hematopoietic stem cells (HSCs) in the bone marrow.1 Once they enter the bloodstream, eosinophils constitute 1–5% of circulating white blood cells in humans,1,2 surviving only a few days in the periphery but extending to weeks during ongoing inflammation. Primarily known for their roles in mediating allergic inflammation, allergy-induced airway remodeling and parasite clearance, eosinophils are known to contribute to the development of inflammatory, allergic, and hyperproliferative disorders.1,3
The accurate identification and quantification of both surface and intracellular eosinophil markers (Table 1) is fundamental for advancing our understanding of eosinophil biology. The availability of well characterized eosinophil marker antibodies enables us to:
Eosinophils express several surface markers, including IL-5Ra (CD125), CCR3 (CD193), Siglec-8 (human) / Siglec-F (mice), as well as CD11b, which is commonly regarded as a pan-granulocyte marker. Characterized by their cytoplasmic granules, which carry a variety of different cargo including cytokines, chemokines, and cytotoxic cationic proteins, eosinophils are also loaded with a number of eosinophil-specific intracellular markers.
| Marker | Cellular Localization | Function |
|---|---|---|
| CCR3 | Cell surface | Constitutively expressed receptor that acts to recruit eosinophils to healthy and diseased target tissues. Also implicated in airway remodeling due to its high expression in airway epithelial cells. |
| C/EBPα, C/EBPβ | Nucleus | These proteins act as developmental regulators that induce eosinophil differentiation. Importantly, they are not expressed in multipotent progenitor cells but are induced during eosinophil lineage commitment. |
| CD11b | Cell surface | This integrin protein is constitutively expressed on eosinophil cell surfaces and becomes upregulated during activation and inflammation. CD11b regulates adhesion to endothelium and extracellular matrix proteins to facilitate tissue recruitment and migration. |
| CD11c | Cell surface | Shows variable expression on eosinophils depending on tissue localization and activation state. CD11c expression is typically low in circulating eosinophils but increases progressively during eosinophil activation and correlates with other activation markers like CD11b and Siglec-F in mice. |
| Eosinophil Cationic Protein (ECP) | Granules (matrix) | Both are cationic granule proteins that have ribonuclease activity, allowing degradation. |
| Eosinophil-Derived Neurotoxin (EDN) | Granules (cytoplasmic) | |
| Eosinophil Peroxidase (EPO) | Granules (matrix) | Also known as EPX. Heme-containing glycoprotein with peroxidase activity. |
| FOG | Nucleus | Known as Friend of Gata (FOG), this transcriptional regulator is highly expressed in hematopoietic progenitors but is quickly downregulated during eosinophil development, making it useful as a negative marker for eosinophil development. |
| GATA-1 | Nucleus | Acts as a lineage-specific marker as well as a developmental marker because eosinophils at different stages of maturation show differential GATA-1 expression. GATA-1 is not eosinophil-specific since it is also expressed in erythroid and megakaryocytic lineages. |
| IL-5Rα | Cell surface | A heterodimeric receptor protein that is expressed on eosinophil cell surfaces from very early developmental stages. |
| Ly6G (mouse) | Cell surface | Normally shows low expression levels on laboratory mouse eosinophils and high expression in wild mice. Ly6G expression on eosinophils is also associated with activation status and can be induced by IL-5 or during fungal allergen challenge. |
| Major Basic Protein (MBP) | Granules (cytoplasmic) | A potent toxin that is implicated in antiparasitic host defense and in allergic and inflammatory conditions. |
| PU.1 | Nucleus | Found in eosinophil progenitors alongside other transcription factors, including GATA-1 and C/EBPα/β, making it useful as part of a marker panel for analyzing eosinophil development. |
| Siglec-8 | Cell surface | As receptor found on human eosinophils (as well as mast cells and basophils), Siglec-8 acts as a late terminal differentiation marker that remains stably expressed despite the activation state. When Siglec-8 binds antibodies or glycan ligands, it can induce apoptosis leading to reactive oxygen species (ROS) production. |
| Siglec-F (mouse) | Cell surface | The murine functional paralog of human Siglec-8, this protein is expressed on mouse eosinophils and alveolar macrophages and plays a similar role in apoptosis. |
Table 1: Key eosinophil marker localization and function
Figure 1: IHC of human pancreas tissue stained with Recombinant Anti-EPX Antibody [EPX/3908R] (A250406).
Figure 2: Flow cytometry analysis of murine peritoneal fluid cell suspension stained with Anti-Ly6g Antibody [RB6-8C5] (Biotin) (A254420).
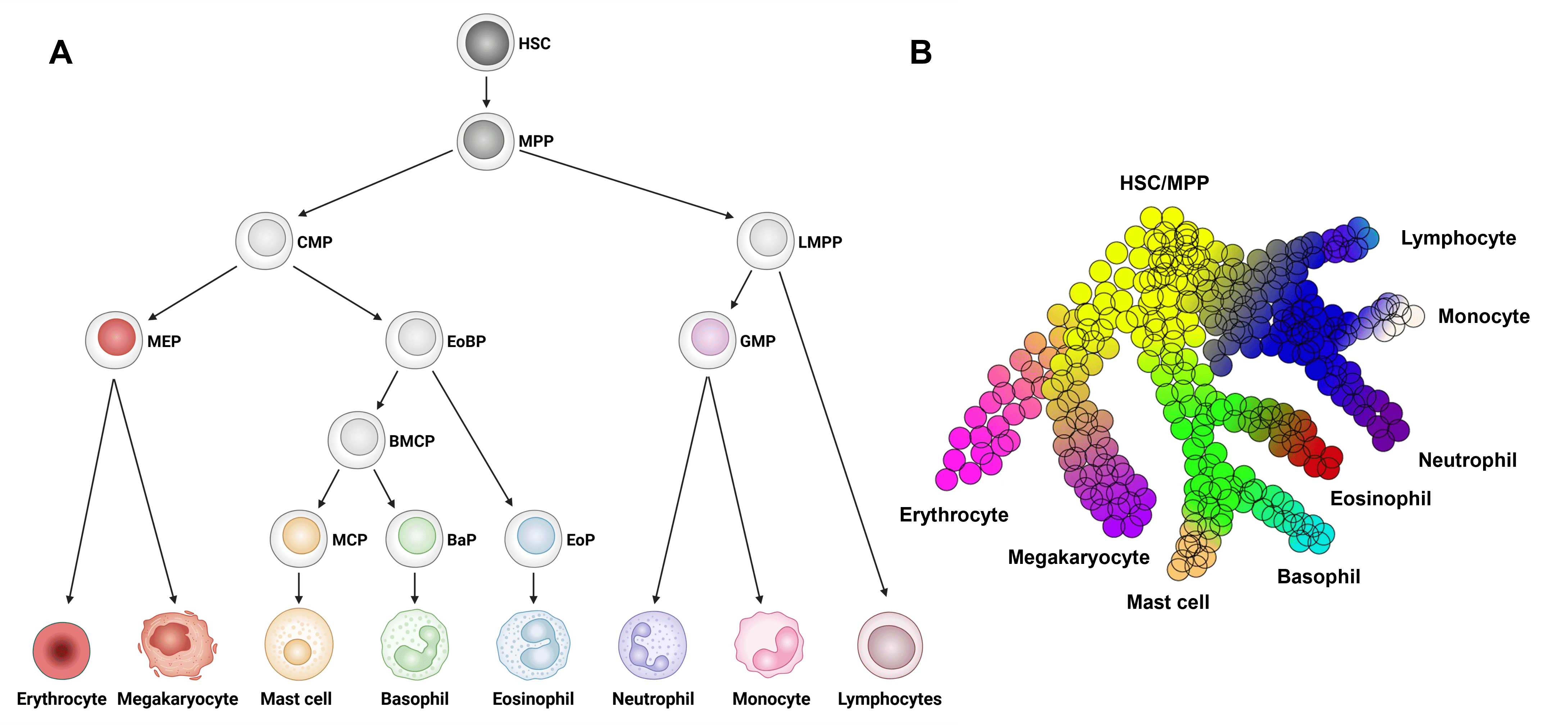
Eosinophils develop from HSCs in the bone marrow via a series of intermediate progenitor stages in a process called granulopoiesis (Figure 3).
Figure 3: Eosinophil development from hematopoietic stem cells. Model of HSC development to eosinophils and other major immune cell types. B, Continuous model of hematopoietic differentiation based on single cell RNAseq analysis. BaP, basophil progenitor; BMCP, basophil/mast cell progenitor; CMP, common myeloid progenitor; EoBP, eosinophil/basophil progenitor; EoP, eosinophil progenitor; GMP, granulocyte/macrophage progenitor; HSC, hematopoietic stem cell; LMPP, lympho-myeloid primed progenitor; MCP, mast cell progenitor; MEP, megakaryocyte/erythrocyte progenitor; MPP, multi-potential progenitor. Edited and reproduced under CC BY 4.0 from 23.
As terminally differentiated cells, eosinophils do not divide or proliferate once they leave the bone marrow. Their development and specialization are controlled through a complex interplay between multiple transcriptional regulators,1 including:
These four regulators can also be used as markers to assess eosinophil lineage and development (see Table 1).6–10
Figure 4: IF of Ramos cells stained with Anti-PU.1 Antibody [PU1/2146] (A250023).
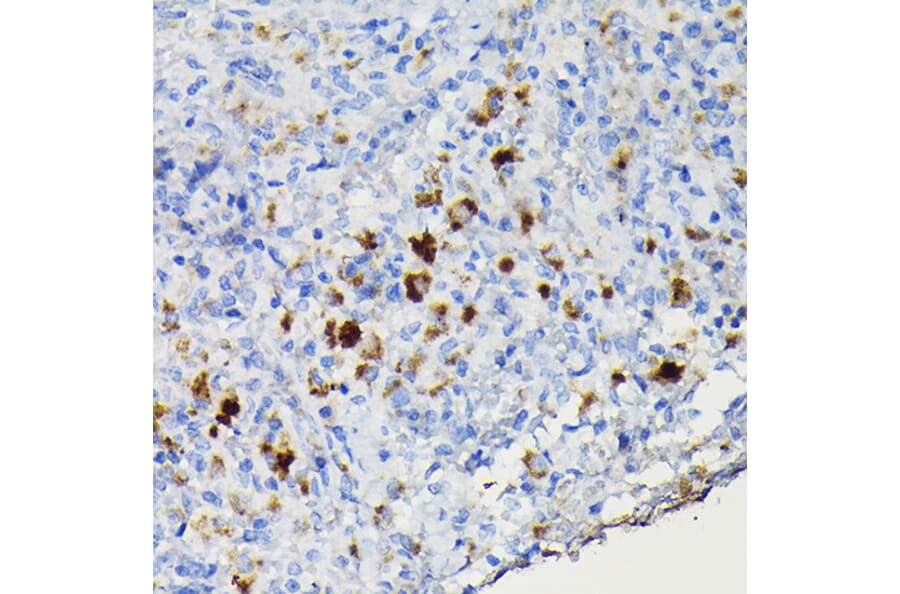
Figure 5: IHC of human spleen tissue stained with Anti-PU.1 Antibody [PU1/2118] (A277809).
As multifunctional leukocytes, eosinophils are implicated in tissue repair and in the activation of proinflammatory responses, particularly in allergic diseases and parasitic helminth infections.3,5,11 Eosinophils act throughout the body; once in circulation, eosinophils migrate to different tissues, including the brain, lower gastrointestinal tract, ovaries, uterus, spleen, and lymph nodes. In addition, during allergic reactions, these cells localize to the lungs, skin, and esophagus.
Following stimulation (e.g. during allergic reactions, pathogen infection), eosinophils are recruited to sites of inflammation where they release their granular contents, including cytotoxic proteins and cytokines, into the extracellular space.1,3 This process, an immunological response called degranulation, is important for destroying invading pathogens, regulating inflammation and for tissue remodeling.3,12 The release of these cargo during degranulation can also cause tissue damage, contributing to the pathophysiology of eosinophil-driven diseases.3,13 More recent, and somewhat controversial, evidence also suggests that these cells may also be play roles in homeostatic and other diseased conditions.3
C-C motif chemokine receptor type 3 (CCR3, also CD193) is highly and constitutively expressed on eosinophil cell surfaces. Like CCR2 in monocytes,14 CCR3 acts to shuttle eosinophils to the appropriate target tissues in both healthy and diseased conditions.15,16 This protein is thought to play a significant role in allergic disease pathogenesis (e.g., asthma, allergic dermatitis, allergic rhinitis).1,15
Importantly, CCR3 expression in airway epithelial cells is upregulated in inflamed asthmatic airways compared to healthy airways.15 The dual expression of CCR3 on eosinophils and epithelial cells allows for a cooperative mechanism whereby epithelial-derived eotaxins recruit CCR3-expressing eosinophils to the airways, while also activating epithelial CCR3 to promote the tissue remodeling process.1,16 Eosinophils and their subsequent inflammatory responses cause significant airway remodeling in different diseases, (e.g., asthma, chronic rhinosinusitis with nasal polyps (CRSwNP)).1,15,16 Therefore, the availability of well characterized anti-CCR3 antibodies permits the study of biological processes involved in airway remodeling to improve disease knowledge and help in the development of novel therapeutic interventions.
Figure 6: Flow cytometry analysis of human peripheral blood stained with Anti-CCR3 Antibody [5E8] (PE) (A86559).
Figure 7: IHC of mouse spleen tissue stained with Anti-CCR3 Antibody (A309001).
Existing as a heterodimer, interleukin-5 receptor alpha (IL-5Rα, also CD125) is expressed on eosinophil cell surfaces during very early stages of cell development. Moreover, eosinophils differentiate when exposed to IL-5. IL-5Rα is, therefore, a useful phenotypic marker for tracking eosinophil development and production,1,2,15 as well as for sorting eosinophil progenitors in vitro.2
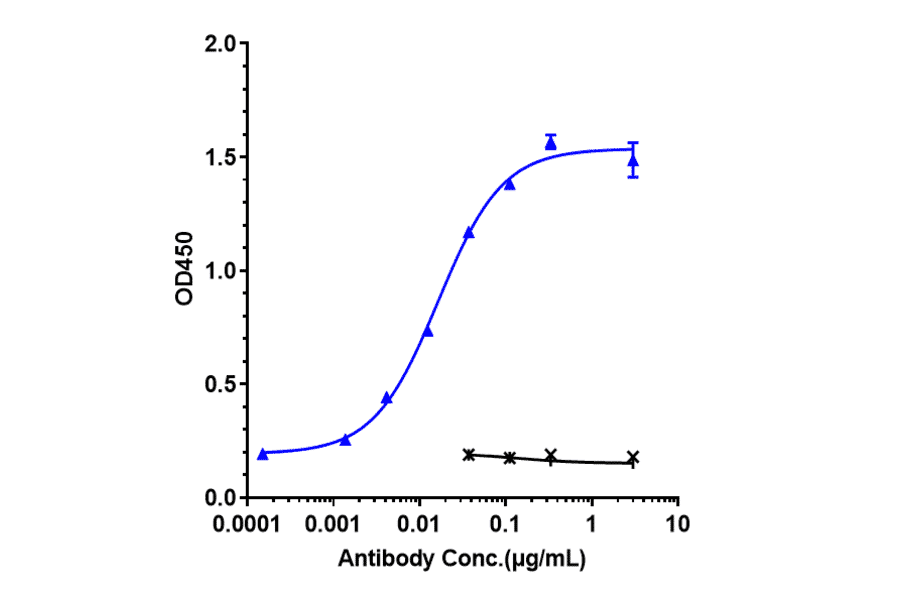
Figure 8: Immobilized recombinant human IL-5R alpha protein bound by Benralizumab Biosimilar - Anti-IL-5RA Antibody - Low endotoxin, Azide free (A323311).
Figure 9: Flow cytometry analysis of Expi293 cells transfected with human IL-5RA (blue) or an irrelevant protein (red) stained with Recombinant Anti-IL-5RA Antibody [DMC393] - BSA and Azide free (A318755).
MBP is a non-enzymatic protein that is found in the crystalline granule core. As a potent toxin, MBP acts to damage both invading parasites and mammalian cells and plays a key role in allergic and inflammatory conditions.1 Like IL-5, MBP expression levels are upregulated in the airways of symptomatic asthma, making it a useful marker to assess the pathogenesis of asthma and other eosinophilic inflammatory conditions.1
Eosinophils are recruited to the lungs following influenza and respiratory syncytial virus (RSV) infection.17,18 ECP (RNase3) and EDN (RNase2), both major granule proteins, function as ribonucleases that degrade the RNA genomes of some invading RNA viruses during host defense.17–19
These proteins may contribute to the observed increase in antiviral activity observed in hypereosinophillic mice that have been shown to clear respiratory syncytial virus (RSV) more effectively than wild-type mice.1,18 Recent studies also suggest that EDN is upregulated during SARS-CoV2 infection.20 As such, ECP and EDN are important markers to assess viral responses and their associated therapeutic interventions. The mechanisms by which beneficial anti-viral and harmful pro-inflammatory responses are regulated are interesting areas that need to be addressed in future research.21
Eosinophils display significant plasticity and heterogeneity in their function, and thus also in their marker expression, influenced by their cellular activation state and tissue microenvironment.
Eosinophils are often categorized as Type 1 or Type 2 based on their response to different cytokines, including interferon gamma (IFN-γ) and IL-4, respectively; type 1 eosinophils are involved in pro-inflammatory responses, while type 2 eosinophils mediate tissue repair and allergies. During activation of type 1 eosinophils, CCR3 can undergo rapid ligand-induced internalization by endocytosis, which transiently reduces cell surface receptor availability. In comparison, type 2 eosinophils retain higher levels of surface expressed CCR3 to maintain chemotaxis to sites of inflammation. Nonetheless, categorization as type 1 or type 2 is not a fixed attribute, and the cytokine profiles and functions of eosinophils can be altered in response to new environmental cues.22
Eosinophil localization also affects marker expression. IL-5Rα is progressively downregulated on the surface of tissue-resident eosinophils compared to cells in the peripheral blood, especially in asthmatic airways where soluble IL-5Rα increase with disease severity.16,21
Other markers, including CD11c and FOG (see Table 1), exhibit dynamic expression patterns in response to external signals, reflecting the functional diversity of eosinophils.
Diagrams created with BioRender.com.
![IHC - Recombinant Anti-EPX Antibody [EPX/3908R] (A250406)](https://cdn.antibodies.com/image/catalog/250/A250406_1.jpg?profile=product_image)
![Flow cytometry - Anti-Ly6g Antibody [RB6-8C5] (Biotin) (A254420)](https://cdn.antibodies.com/image/catalog/254/A254420_1.jpg?profile=product_image)
![IF - Anti-PU.1 Antibody [PU1/2146] (A250023)](https://cdn.antibodies.com/image/catalog/250/A250023_7.jpg?profile=product_image)
![IHC - Anti-PU.1 Antibody [PU1/2118] (A277809)](https://cdn.antibodies.com/image/catalog/277/A277809_4.jpg?profile=product_image)
![Flow cytometry - Anti-CCR3 Antibody [5E8] (PE) (A86559)](https://cdn.antibodies.com/image/catalog/86/A86559_725.tif?profile=product_image)
![Flow cytometry - Recombinant Anti-IL-5RA Antibody [DMC393] - BSA and Azide free (A318755)](https://cdn.antibodies.com/image/catalog/318/A318755_1.jpg?profile=product_image)